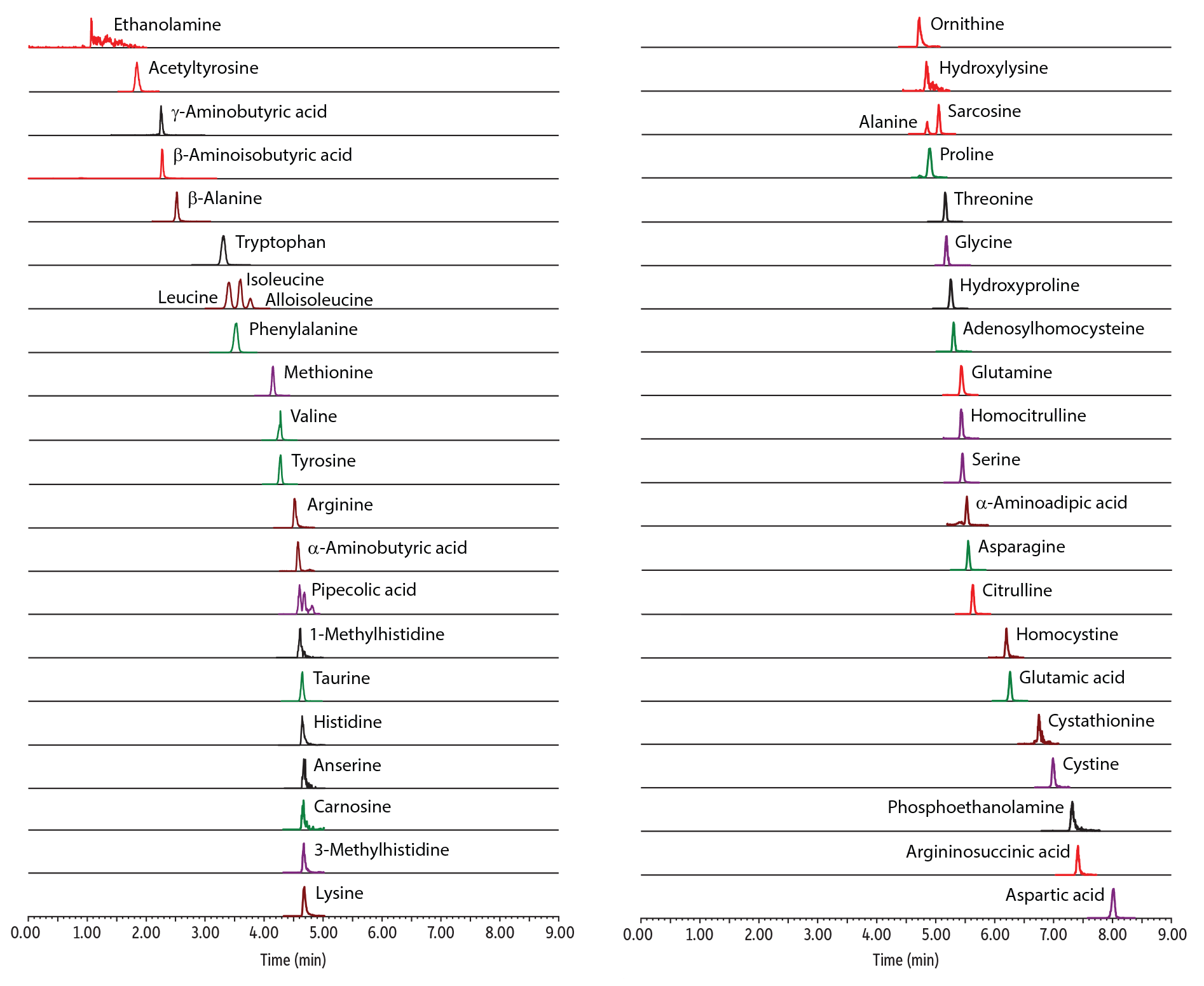
13-Minute, Comprehensive, Direct LC-MS/MS Analysis of Amino Acids in Plasma
Abstract
A fast, direct method for LC-MS/MS analysis of amino acids in plasma was developed during the course of this study. Using a simple sample preparation procedure without derivatization and a hybrid retention mode Raptor Polar X column, 45 amino acids were simultaneously analyzed in a 13-minute run. Good chromatographic results, including the separation of critical isobaric compounds, were achieved and acceptable accuracy, precision, and linearity results were obtained.
Introduction
Amino acids are critical molecules for biological function as they are the fundamental building blocks of peptides and proteins and also serve as intermediates in various metabolic pathways, such as the urea cycle and the citric acid cycle. Amino acid profiling in human plasma is an important tool for the diagnosis of metabolic disorders, especially for inborn errors of metabolism. A fast and accurate large-panel analysis of amino acids in plasma is critical not only for disorder identification but also for continued monitoring and assessment of the nutritional status of patients and the effectiveness of treatment plans.
Traditionally, amino acids have been analyzed by either post-column or precolumn derivatization methodologies; however, these approaches have significant limitations. The major shortcoming of post-column derivatization methods is that chromatographic cycling times are typically quite long, which limits sample throughput capacity. Precolumn derivatization methods often have poor chromatographic separation, which can reduce the number of amino acids that can be included in comprehensive amino acid panels. Direct LC-MS/MS analysis of amino acids in plasma is an attractive alternative because no costly and time-consuming derivatization step is necessary, but, historically, there has been a relative lack of direct analysis applications due to poor chromatographic retention and insufficient detection sensitivity.
With the recent advancements in hydrophilic-interaction chromatography (HILIC) LC columns and modern MS instrumentation, it is now possible to analyze underivatized amino acids with simplicity and consistency. In this study, a straightforward workflow was developed and implemented for accurate quantification of a large panel of 45 amino acids in human plasma. A fast chromatographic analysis (13 minutes) was achieved using a Raptor Polar X column. This column was selected because the unique hybrid (HILIC and ion exchange) stationary phase has previously been demonstrated to adequately retain and effectively separate a wide range of amino acid chemistries without derivatization [1].
Experimental
Standard and Control Plasma Materials
Two standard mixture solutions (A6282 and A6407) containing a total of 28 amino acids and 17 solid standards were obtained from Sigma-Aldrich. A stock standard solution containing 45 amino acids was prepared in phosphate buffered saline (1x PBS) solution at 250-500 µmol/L.
Internal standards were used to compensate for plasma matrix effects. The internal standard (IS) working solution was prepared by mixing a stock solution of 17 isotopically labeled amino acids (MSK-A2-S, Cambridge Isotope Laboratories) with 12 individual deuterium isotopes of α-aminobutyric acid, β-aminoisobutyric acid, γ-aminobutyric acid, β-alanine, N-acetyltyrosine, asparagine, ethanolamine, glutamine, homocystine, 1-methylhistidine, pipecolic acid, and taurine (CDN Isotopes) in water at 125-250 µmol/L.
MassChrom amino acid analysis plasma control level I (0471), II (0472), and III (0473) samples were obtained from Chromsystems.
Calibration Standard Preparation
For this LC-MS/MS analysis of amino acids in plasma, calibration standards were prepared across a range of 1-500 µmol/L in 1x PBS solution. A 25 µL aliquot of calibration standard was mixed with 2.5 µL of 30% sulfosalicylic acid solution, 2 µL of internal standard working solution, and 225 µL of mobile phase B, acetonitrile:water (90:10), 0.5% formic acid and 1 mM ammonium formate, for injection and LC-MS/MS analysis. Exploratory studies demonstrated that the chromatographic retention of three analytes, β-aminoisobutyric acid, γ-aminobutyric acid, and β-alanine, is sensitive to the salt concentration in final injection solution. Therefore, the standards were prepared in PBS in order to match the salt condition in plasma samples and ensure consistent retention times for these three compounds between the standard and sample solutions.
Sample Preparation
Preliminary testing established that overall detection sensitivity was greatly improved by performing a protein precipitation step using 30% sulfosalicylic acid. Therefore, a 50 µL aliquot of control plasma was mixed with 5 µL of 30% sulfosalicylic acid solution. Following centrifugation at 4200 rpm for 10 minutes to pellet the precipitated material, a 27.5 µL aliquot of clear supernatant was mixed with 2 µL of internal standard working solution and 225 µL of mobile phase B prior to LC-MS/MS analysis.
Chromatographic Method
The chromatographic conditions used on a Waters ACQUITY UPLC coupled with a Xevo TQ-S mass spectrometer for this LC-MS/MS analysis of amino acids in plasma are detailed below. The ion transitions and internal standards used for each analyte are provided in Table I.
During method development experiments, several chromatographic parameters were determined to be critical for successful analysis. The column temperature of 35 °C was needed to have a stable, well-formed γ-aminobutyric acid peak. In addition, using a guard column and starting the analysis with 96%B mobile phase provided the most consistent peak shape for ethanolamine. Also, while all analytes elute by eight minutes, adding a one-minute wash with a highly aqueous mobile phase was found to be beneficial in reducing carryover for aspartic acid, argininosuccinic acid, anserine, carnosine, adenosylhomocysteine, homocystine, cystathionine, citrulline, 3-methylhistidine, 1-methylhistidine, and phosphoethanolamine. Finally, sensitivity issues were observed for glycine, and the shallow gradient used here gave better results than the steeper gradients that were also assessed.
| Column: | Raptor Polar X (2.7 µm, 100 mm x 2.1 mm ID [cat.# 9311A12]) | ||||||||||||||||
| Guard column: | Raptor Polar X EXP guard column cartridge (2.7 µm, 5 mm x 2.1 mm [cat.# 9311A0252]) | ||||||||||||||||
| Column temp.: | 35 °C | ||||||||||||||||
| Injection volume: | 5 µL | ||||||||||||||||
| Mobile phase A: | Water, 0.5% formic acid and 1 mM ammonium formate | ||||||||||||||||
| Mobile phase B: | Acetonitrile:water (90:10), 0.5% formic acid and 1 mM ammonium formate | ||||||||||||||||
|
|||||||||||||||||
| Flow rate: | 0.3 mL/min | ||||||||||||||||
| Ion mode: | Positive ESI | ||||||||||||||||
| Mode: | Scheduled MRM |
Table I: Ion Transitions and Internal Standards for LC-MS/MS Analysis of Amino Acids in Plasma.
|
Peak Identification |
Retention Time (min) |
Precursor Ion |
Product Ion |
Internal Standard |
|
Ethanolamine |
1.06 |
62.0 |
44.1 |
Ethanolamine-d4 |
|
Acetyltyrosine |
1.83 |
224.1 |
136.1 |
Acetyltyrosine-d3 |
|
γ-Aminobutyric acid |
2.25 |
104.0 |
69.0 |
γ-Aminobutyric acid-d4 |
|
β-Aminoisobutyric acid |
2.27 |
104.1 |
30.0 |
β-Aminoisobutyric acid-d3 |
|
β-Alanine |
2.52 |
90.0 |
30.1 |
ß-Alanine-d4 |
|
Tryptophan |
3.31 |
205.1 |
146.1 |
ß-Alanine-d4 |
|
Leucine |
3.40 |
132.1 |
86.1 |
Leucine 13C6 15N |
|
Phenylalanine |
3.53 |
166.1 |
120.1 |
Phenylalanine 13C9 15N |
|
Isoleucine |
3.60 |
132.1 |
86.1 |
Isoleucine 13C6 15N |
|
Alloisoleucine |
3.77 |
132.1 |
86.1 |
Methionine 13C5 15N |
|
Methionine |
4.15 |
150.1 |
104.1 |
Methionine 13C5 15N |
|
Tyrosine |
4.27 |
182.1 |
136.1 |
Tyrosine 13C9 15N |
|
Valine |
4.27 |
118.1 |
72.1 |
Valine 13C5 15N |
|
Arginine |
4.52 |
175.2 |
70.1 |
Arginine 13C6 15N4 |
|
α-Aminobutyric acid |
4.57 |
104.0 |
58.1 |
α-Aminobutyric acid-d2 |
|
Pipecolic acid |
4.60 |
130.0 |
84.1 |
Pipecolic acid-d9 |
|
1-Methylhistidine |
4.62 |
170.1 |
124.1 |
1-Methylhistidine-d3 |
|
Histidine |
4.65 |
156.1 |
110.2 |
Histidine 13C6 15N3 |
|
Taurine |
4.65 |
126.1 |
108.1 |
Taurine-d4 |
|
Anserine |
4.67 |
241.2 |
109.1 |
Histidine 13C6 15N3 |
|
Carnosine |
4.67 |
227.2 |
110.1 |
Histidine 13C6 15N3 |
|
Lysine |
4.68 |
147.1 |
84.1 |
Lysine 13C6 15N2 |
|
3-Methylhistidine |
4.68 |
170.0 |
126.0 |
Histidine 13C6 15N3 |
|
Ornithine |
4.72 |
133.1 |
70.0 |
Lysine 13C6 15N2 |
|
Hydroxylysine |
4.84 |
163.1 |
128.1 |
Threonine 13C4 15N |
|
Alanine |
4.86 |
90.0 |
44.1 |
Alanine 13C3 15N |
|
Proline |
4.89 |
116.1 |
70.1 |
Proline 13C5 15N |
|
Sarcosine |
5.05 |
90.0 |
44.1 |
Threonine 13C4 15N |
|
Threonine |
5.15 |
120.1 |
74.1 |
Threonine 13C4 15N |
|
Glycine |
5.19 |
76.1 |
30.1 |
Threonine 13C4 15N |
|
Hydroxyproline |
5.26 |
132.1 |
86.1 |
Threonine 13C4 15N |
|
Adenosylhomocysteine |
5.30 |
385.0 |
133.7 |
Threonine 13C4 15N |
|
Glutamine |
5.43 |
147.1 |
84.1 |
Glutamine-d5 |
|
Homocitrulline |
5.43 |
190.1 |
84.1 |
Glutamine-d5 |
|
Serine |
5.46 |
106.1 |
60.1 |
Serine 13C3 15N |
|
α-Aminoadipic acid |
5.53 |
162.1 |
98.1 |
Asparagine-d3 |
|
Asparagine |
5.54 |
133.1 |
74.1 |
Asparagine-d3 |
|
Citrulline |
5.61 |
176.1 |
159.1 |
Asparagine-d3 |
|
Homocystine |
6.20 |
269.1 |
136.0 |
Homocystine-d8 |
|
Glutamic acid |
6.26 |
148.1 |
84.1 |
Glutamic Acid 13C5 15N |
|
Cystathionine |
6.75 |
223.1 |
134.0 |
Glutamic Acid 13C5 15N |
|
Cystine |
6.99 |
241.1 |
152.0 |
Cystine 13C6 15N2 |
|
Phosphoethanolamine |
7.32 |
142.0 |
44.1 |
Cystine 13C6 15N2 |
|
Argininosuccinic acid |
7.41 |
291.1 |
70.1 |
Cystine 13C6 15N2 |
|
Aspartic acid |
8.00 |
134.1 |
74.1 |
Aspartic Acid 13C4 15N |
Results and Discussion
Chromatographic Performance
A fast, comprehensive method was established for the direct LC-MS/MS analysis of amino acids in plasma. In total, 45 amino acids were simultaneously analyzed on a Raptor Polar X column with a 13-minute total cycle time (Figure 1). As shown in Figure 2, the selectivity of the Raptor Polar X column provided proper chromatographic separation of the isobaric compounds leucine/isoleucine/alloisoleucine and alanine/sarcosine, which is essential for accurate identification and quantification because these compounds cannot be distinguished by MS alone.
Alloisoleucine is of particular concern because it is an important biomarker for diagnosing maple syrup urine disease, so chromatographic separation from its isobars (leucine and isoleucine) is clinically essential. During method development, the balance of ammonium formate and formic acid concentration in the mobile phase proved to be critical for this separation. Using the mobile phases that were established during method development and starting the run with a two-minute isocratic period, as presented here, proved to be essential for the separation of alloisoleucine, isoleucine, and leucine. In addition, it was observed that isomer resolution can vary among LC instruments, so additional gradient changes and/or adjustments for differences in instrumental dead volume may be necessary when transferring the method between instruments.
Linearity
Various calibration ranges were determined individually for each amino acid based on differences in MS detection sensitivity and diagnostic suitability for all 45 analytes (Table II). All compounds showed acceptable linearity with r2 value >0.990 and deviations <20% using quadratic regression (1/x weighted) calibration curves. While the highest quantifiable concentration of 500 µmol/L is suitable for most of the analytes, it is necessary to dilute the plasma sample for the analysis of glutamine and alanine into this range due to their relatively higher physiological concentration of >500 µmol/L. In addition, due to poor ionization, the LOQ of glycine is much higher than the other amino acid compounds, and the detection sensitivity could vary on different MS systems. Nevertheless, as the physiological concentration of glycine is relatively higher, this LC-MS/MS method is still suitable for glycine quantification in plasma.
Accuracy and Precision
Three control plasma samples containing low to high concentrations of all 45 analytes were used to evaluate method accuracy and precision. The acceptance criteria for method accuracy is that the measured concentrations fall within the labeled concentration ranges for each analyte. A total of three batches were analyzed on different days. Table II shows the accuracy and precision results averaged across all three batches (n=9). Method accuracy criteria were met, as demonstrated by the concentration values falling within the nominal ranges for all 45 analytes. In addition, %RSD values were <20%, which indicates that method precision was also acceptable.
Figure 1: Control Plasma Level 2
Figure 2: Control Plasma Level 1–Chromatographic Separation of Critical Isobaric Groups (Leucine/Isoleucine/Alloisoleucine and Alanine/Sarcosine).
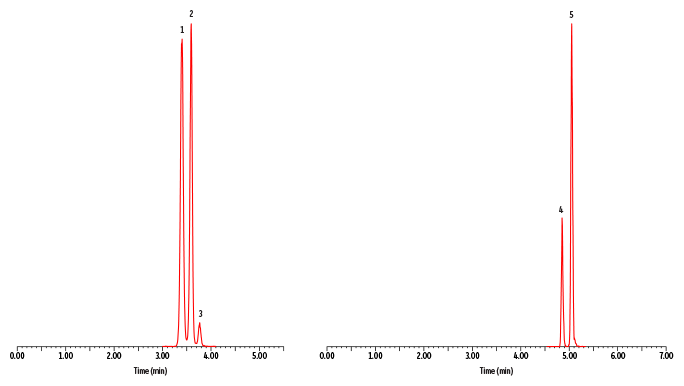
| Peaks | tR (min) | Precursor Ion | Product Ion | |
|---|---|---|---|---|
| 1. | Leucine | 3.40 | 132.1 | 86.1 |
| 2. | Isoleucine | 3.59 | 132.1 | 86.1 |
| 3. | Alloisoleucine | 3.77 | 132.1 | 86.1 |
| 4. | Alanine | 4.85 | 90.0 | 44.1 |
| 5. | Sarcosine | 5.04 | 90.0 | 44.1 |
Table II: Accuracy and Precision Results for LC-MS/MS Analysis of Amino Acids in Plasma.
| Control Level 1 | Control Level 2 | Control Level 3 | |||||
|
Analyte |
Linearity Range (µmol/L) |
Nominal Conc. Range (µmol/L) |
Average Conc. (µmol/L) (%RSD) |
Nominal Conc. Range (µmol/L) |
Average Conc. (µmol/L) (%RSD) |
Nominal Conc. Range (µmol/L) |
Average Conc. (µmol/L) (%RSD) |
|
Acetyltyrosine |
0.5 - 250 |
4.10 - 6.15 |
4.84 (9.76) |
32.5 - 48.8 |
40.5 (9.76) |
58.5 - 87.7 |
73.9 (15.1) |
|
Adenosylhomocysteine |
2.5 - 250 |
2.77 - 4.16 |
3.16 (15.0) |
17.8 - 26.7 |
20.4 (15.4) |
32.5 - 48.8 |
42.9 (9.98) |
|
Alanine |
5 - 500 |
159 - 239 |
191 (15.0) |
524 - 786 |
655* (20.0) |
890 - 1334 |
979* (19.4) |
|
β-Alanine |
1 - 500 |
10.4 - 15.5 |
10.8 (3.13) |
42.7 - 64.0 |
47.9 (3.12) |
74.0 - 111 |
81.1 (3.01) |
|
α-Aminoadipic acid |
1 - 500 |
4.98 - 7.46 |
5.74 (7.53) |
9.92 - 14.9 |
11.2 (6.75) |
14.7 - 22.1 |
17.9 (11.7) |
|
α-Aminobutyric acid |
1 - 500 |
3.69 - 5.54 |
4.47 (11.4) |
39.5 - 59.2 |
52.1 (10.9) |
73.6 - 110 |
97.7 (7.43) |
|
β-Aminoisobutyric acid |
1 - 500 |
3.82 - 5.73 |
4.40 (9.85) |
17.0 - 25.5 |
19.2 (6.10) |
29.7 - 44.5 |
35.3 (7.75) |
|
γ-Aminobutyric acid |
1 - 500 |
3.79 - 5.69 |
4.75 (5.35) |
7.98 - 12.0 |
9.60 (3.45) |
12.0 - 17.9 |
14.1 (5.60) |
|
Anserine |
1 - 500 |
3.85 - 5.77 |
4.71 (12.5) |
7.65 - 11.5 |
10.8 (5.82) |
11.5 - 17.3 |
15.9 (7.71) |
|
Arginine |
1 - 500 |
8.63 - 12.9 |
9.55 (11.5) |
124 - 186 |
133 (3.60) |
237 - 356 |
257 (3.32) |
|
Argininosuccinic acid |
0.5 - 250 |
2.35 - 3.53 |
2.96 (13.3) |
59.7 - 89.6 |
72.3 (11.1) |
115 - 172 |
141 (7.99) |
|
Asparagine |
1 - 500 |
21.4 - 32.1 |
31.3 (2.46) |
97.0 - 145 |
143 (1.39) |
173 - 259 |
250 (2.22) |
|
Aspartic acid |
1 - 500 |
18.5 - 27.7 |
19.2 (1.83) |
101 - 152 |
105 (3.54) |
182 - 274 |
189 (2.89) |
|
Carnosine |
1 - 500 |
7.69 - 11.5 |
10.3 (9.66) |
18.9 - 28.3 |
23.7 (10.1) |
29.2 - 43.8 |
36.6 (10.2) |
|
Citrulline |
5 - 500 |
12.7 - 19.1 |
15.6 (13.9) |
83.8 - 126 |
88.8 (4.17) |
153 - 230 |
161 (1.63) |
|
Cystathionine |
1 - 500 |
4.86 - 7.29 |
5.76 (6.47) |
20.1 - 30.1 |
23.1 (8.07) |
35.3 - 52.9 |
40.7 (9.63) |
|
Cystine |
0.5 - 250 |
8.54 - 12.8 |
10.3 (9.50) |
62.7 - 94.0 |
78.8 (6.33) |
116 - 174 |
144 (9.05) |
|
Ethanolamine |
5 - 250 |
12.3 - 18.4 |
15.2 (11.1) |
98.5 - 148 |
125 (11.3) |
182 - 273 |
206 (3.53) |
|
Glutamic acid |
1 - 500 |
45.8 - 68.7 |
53.7 (6.16) |
304 - 456 |
335 (8.12) |
553 - 829 |
593* (8.49) |
|
Glutamine |
1 - 500 |
286 - 429 |
371 (3.58) |
748 - 1122 |
975* (5.46) |
1209 - 1814 |
1587* (8.40) |
|
Glycine |
25 - 500 |
154 - 231 |
177 (10.2) |
517 - 775 |
613* (7.54) |
864 - 1295 |
995* (10.9) |
|
Histidine |
1 - 500 |
35.9 - 53.9 |
47.6 (5.27) |
121 - 182 |
157 (7.27) |
210 - 316 |
266 (4.35) |
|
Homocitrulline |
2.5 - 250 |
7.88 - 11.8 |
9.87 (2.05) |
22.6 - 33.9 |
31.3 (3.83) |
37.1 - 55.6 |
53.7 (2.69) |
|
Homocystine |
1 - 500 |
3.29 - 4.93 |
3.60 (9.17) |
10.5 - 15.8 |
12.8 (5.69) |
18.0 - 27.0 |
22.0 (4.71) |
|
Hydroxylysine |
5 - 500 |
3.97 - 5.96 |
5.50 (7.60) |
14.3 - 21.4 |
16.58 (11.2) |
24.3 - 36.5 |
28.4 (14.2) |
|
Hydroxyproline |
5 - 500 |
7.94 - 11.9 |
10.1 (14.2) |
72.8 - 109 |
97.52 (8.40) |
136 - 204 |
198 (5.02) |
|
Allo-isoleucine |
0.5 - 250 |
2.86 - 4.29 |
3.55 (3.12) |
65.5 - 98.2 |
79.15 (4.56) |
128 - 192 |
150 (5.87) |
|
Isoleucine |
5 - 500 |
18.4 - 27.6 |
20.1 (3.12) |
143 - 215 |
164.41 (3.01) |
260 - 389 |
282 (2.69) |
|
Leucine |
5 - 500 |
40.2 - 60.3 |
43.3 (2.57) |
239 - 358 |
288.87 (4.37) |
444 - 666 |
512* (4.67) |
|
Lysine |
1 - 500 |
21.9 - 32.9 |
28.2 (5.79) |
231 - 346 |
315.89 (4.13) |
436 - 655 |
621* (4.61) |
|
Methionine |
1 - 500 |
6.80 - 10.2 |
7.46 (5.29) |
64.8 - 97.1 |
72.02 (3.83) |
121 - 181 |
139 (2.32) |
|
1-Methylhistidine |
1 - 500 |
2.06 - 3.08 |
2.52 (8.18) |
6.17 - 9.25 |
7.12 (9.84) |
10.3 - 15.4 |
12.4 (9.84) |
|
3-Methylhistidine |
5 - 500 |
5.82 - 8.73 |
8.03 (8.31) |
36.2 - 54.3 |
43.81 (10.2) |
66.3 - 99.5 |
85.1 (4.10) |
|
Ornithine |
5 - 500 |
13.6 - 20.4 |
15.2 (3.13) |
161 - 242 |
168.76 (4.35) |
301 - 452 |
322 (5.76) |
|
Phenylalanine |
1 - 500 |
32.2 - 48.3 |
36.5 (3.38) |
248 - 373 |
297.96 (1.80) |
461 - 692 |
547* (4.31) |
|
Phosphoethanolamine |
5 - 500 |
8.53 - 12.8 |
9.64 (12.2) |
57.4 - 86.1 |
64.32 (9.09) |
105 - 158 |
127 (9.93) |
|
Pipecolic acid |
0.5 - 250 |
1.62 - 2.43 |
1.85 (9.98) |
13.4 - 20.0 |
17.25 (8.08) |
24.6 - 36.9 |
31.0 (7.45) |
|
Proline |
1 - 500 |
50.7 - 76.1 |
58.6 (4.05) |
268 - 403 |
322.45 (3.46) |
482 - 722 |
578* (4.99) |
|
Sarcosine |
1 - 500 |
3.29 - 4.94 |
4.00 (7.35) |
13.5 - 20.3 |
19.59 (6.51) |
23.7 - 35.5 |
34.8 (4.19) |
|
Serine |
5 - 500 |
102 - 153 |
126 (3.65) |
295 - 443 |
383.86 (6.51) |
484 - 726 |
627* (5.89) |
|
Taurine |
5 - 500 |
14.7 - 22.1 |
18.2 (6.28) |
173 - 260 |
234.74 (3.51) |
325 - 488 |
433 (6.26) |
|
Threonine |
1 - 500 |
36.7 - 55.1 |
42.6 (2.76) |
203 - 304 |
246.86 (7.01) |
366 - 549 |
464 (4.16) |
|
Tryptophan |
1 - 500 |
13.5 - 20.2 |
16.9 (11.5) |
88.4 - 133 |
107.52 (7.72) |
158 - 237 |
188 (10.3) |
|
Tyrosine |
1 - 500 |
22.3 - 33.5 |
25.4 (5.35) |
129 - 194 |
148.82 (2.58) |
232 - 348 |
264 (3.40) |
|
Valine |
1 - 500 |
74.4 - 112 |
91.3 (6.73) |
256 - 384 |
318.69 (7.67) |
442 - 664 |
532* (9.56) |
*Concentration is outside of the calibration range.
Conclusion
This comprehensive method for the direct LC-MS/MS analysis of amino acids in plasma demonstrated that 45 amino acids could be simultaneously measured using a simple sample preparation procedure and a Raptor Polar X column. Acceptable accuracy, precision, and linearity results were obtained without derivatization in a fast, 13-minute run, making this approach suitable for high-throughput analysis of amino acids.
References
- Restek Corporation, Fast, direct analysis of underivatized amino acids in infant formula. 2021, https://www.restek.com/articles/fast-direct-analysis-of-underivatized-amino-acids-in-infant-formula/ (accessed July 23, 2021).
This method has been developed for research use only; it is not suitable for use in diagnostic procedures without further evaluation.