Faster Semivolatiles Analysis with a Scaled-Down Method and GC Accelerator Kit
Analysis times for semivolatile compounds can limit sample throughput and decrease overall lab productivity. You can significantly speed up methods on your existing GC-MS by using a properly scaled-down column and appropriately adjusted instrument conditions. The challenge with this approach is that the new conditions can result in oven ramp rates that exceed the performance of standard GC ovens, especially 120 V ovens. However, by using a GC Accelerator oven insert kit, Agilent 6890 and 7890 GCs can meet aggressive scaled-down oven programs, producing the same chromatographic separations in much less time.
In this technical article, we will walk through an example of scaling down EPA Method 8270 and using a GC Accelerator kit to reduce cycle time. Significantly faster semivolatiles analysis times were achieved, providing an opportunity for labs to increase sample throughput or process more rush samples.
Scaling Down Your Semivolatiles Column
To achieve the same chromatographic results in less time, the first step is to take the traditional column for semivolatiles analysis, a 30 m long, 0.25 mm ID, 0.25 µm df arylene-type column (e.g., an Rxi-5Sil MS column) and scale it down to a more efficient format that retains the same number of theoretical plates as the original column. A 20 m long, 0.15 mm ID, 0.15 µm df Rxi-5Sil MS column (cat.# 43816) fits these requirements. The proportionally narrower ID and thinner film will improve efficiency (i.e., give narrower peaks) and provide the same selectivity by keeping the phase ratio (β) constant. Higher efficiency results in more theoretical plates per meter of column, so by shortening the column to 20 m, a very similar total number of plates is achieved. As summarized in Table I, these changes result in the same degree of separation that is obtained on the traditional column format.
Table I: Properly scaled-down columns provide the same separation in less time.
| Column Formats | ||||
| Traditional | Scaled-down | Effect | ||
| Part Number | 13623 | 43816 | ||
| ID (mm) | 0.25 | > | 0.15 | Narrower ID creates greater efficiency |
| Film Thickness, df (µm) | 0.25 | > | 0.15 | Thinner film creates greater efficiency |
| Relative efficiency (plates/m) compared to traditional format | 100% | < | 141% | The scaled-down column is more efficient and will produce narrower peaks |
| Phase Ratio (β) | 250 | = | 250 | Equivalent β will maintain the same selectivity between columns |
| Length (m) | 30 | > | 20 | Shortening the scaled-down column will create a similar total number of plates |
| Relative Total Plates compared to traditional format | 100% | ≈ | 94% | Very similar total number of plates means the same degree of separation in less time |
Scaling Down Method Conditions with the EZGC Method Translator
A scaled-down column requires a scaled-down method to ensure that compound elution temperatures will be the same so that equivalent separations will be achieved in a faster analysis time. Correctly translating the scaled-down column flow rates and oven temperature ramp rates can be a challenge, but this task is made simple by using Restek’s EZGC method translator. This free, online tool calculates properly translated method conditions based on your original column and method in seconds. Just input your current column dimensions, flow parameters, and oven program, then define the new column format you’d like to use, and the software will do the rest.
To set up a comparison of traditional vs. scaled-down, faster semivolatiles analysis, Figure 1 shows a chromatogram generated using EPA 8270D method conditions, split injection, and the traditional column format. In this case, the analysis time has already been shortened by a few minutes from the typical splitless injection approach by the successful adoption of a split injection.
Figure 1: Split analysis of semivolatiles on a traditional 30 m, 0.25 mm ID, 0.25 µm df Rxi-5Sil MS column.
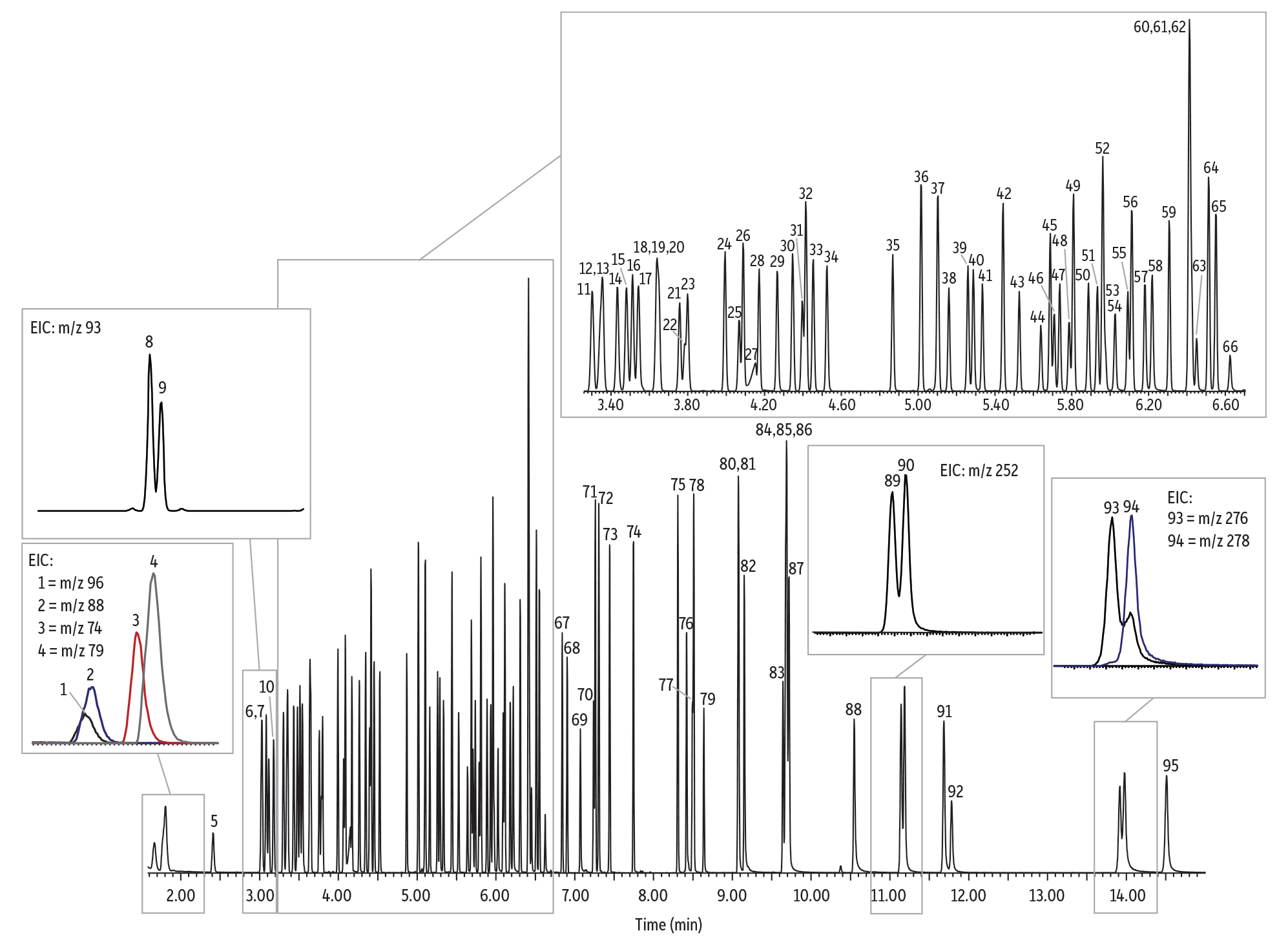
| Column | Rxi-5Sil MS, 30 m, 0.25 mm ID, 0.25 µm (cat.# 13623) |
|---|---|
| Standard/Sample | 8270 MegaMix (cat.# 31850) |
| 8270 Benzidines mix (cat.# 31852) | |
| Benzoic acid (cat.# 31879) | |
| 1,4-Dioxane (cat.# 31853) | |
| Revised B/N surrogate mix (cat.# 31888) | |
| Acid surrogate mix (4/89 SOW) (cat.# 31063) | |
| Revised SV internal standard mix (cat.# 31886) | |
| Diluent: | Methylene chloride |
| Conc.: | 40 µg/mL (IS/SS 20 µg/mL) |
| Injection | |
| Inj. Vol.: | 1 µL split (split ratio 10:1) |
| Liner: | Premium 4 mm Precision liner w/wool (cat.# 23305) |
| Inj. Temp.: | 270 °C |
| Split Vent Flow Rate: | 12 mL/min. |
| Oven | |
| Oven Temp.: | 70 °C (hold 1 min) to 285 °C at 28 °C/min to 305 °C at 3 °C/min to 320 °C at 30 °C/min (hold 1 min) |
| Carrier Gas | He, constant flow |
| Flow Rate: | 1.2 mL/min |
| Detector | MS |
|---|---|
| Mode: | Scan |
| Transfer Line Temp.: | 280 °C |
| Analyzer Type: | Quadrupole |
| Source Temp.: | 270 °C |
| Quad Temp.: | 150 °C |
| Electron Energy: | 70 eV |
| Solvent Delay Time: | 1.3 min |
| Tune Type: | DFTPP |
| Ionization Mode: | EI |
| Scan Range: | 35-550 amu |
| Scan Rate: | 5.36 scans/sec |
| Instrument | Agilent 7890A GC & 5975C MSD |
Figure 2 shows how the split injection semivolatiles analysis illustrated in Figure 1 can be easily translated to the scaled-down column format using the EZGC method translator. Split injection is particularly important with the scaled-down analysis to avoid overloading the column and to transfer a sample band narrow enough to be compatible with the very narrow peaks that the more efficient, scaled-down column produces. Because the narrower peaks are also taller, there is the additional benefit of enough sensitivity to still meet the low detection limits commonly required for semivolatiles analysis. In the scaled-down example shown later in this article, the split ratio was successfully increased from 10:1 to 20:1.
Note that in the examples presented here, the final oven ramp rates and final hold times may differ from the method translator output in Figure 2. The final oven ramp rate shown in the original method of the EZGC method translator example was lowered to 20 °C/min (compared to 30 °C/min Figure 1) in order to represent a rate that is attainable by analysts using Agilent 6890 and 7890 120 V GC ovens. Because the last compound elutes before the final oven ramp, these differences do not affect the chromatographic comparison. Final hold times also do not affect the chromatographic separations and are generally defined in analytical methods.
Figure 2: Use the EZGC method translator to move from a traditional column format to a scaled-down column for faster semivolatiles analysis: (a) Fields to input original method conditions; (b) field to input new column dimensions; (c) calculated scaled method conditions, with ramp rates that pose a problem for certain GCs but that can be resolved with the GC Accelerator kit; (d) calculated analysis time changes.
With the scaled-down column and translated conditions defined, you are nearly ready to try the new method in the lab, but, first, you need to review the oven ramp rates and ensure your GC oven can meet them. In this example, the scaled-down oven program has aggressive ramp rates that some GC ovens can’t consistently achieve. As shown in Table II, 120 V ovens would not be able to meet the new scaled-down oven program reliably, which would result in inconsistent retention times for compounds eluting during that ramp. Also, from the table, it is clear that this problem is easily resolved by the use of the GC Accelerator kit.
Table II: Maximum oven ramp rates for Agilent 6890 and 7890 GC ovens with and without the GC Accelerator kit.
| Temperature Range (°C) | 120 V Oven Ramp Rate (˚C/min) | >200 V Oven Ramp Rate (˚C/min) | ||
| Without GC Accelerator Kit | With GC Accelerator Kit | Without GC Accelerator Kit | With GC Accelerator Kit | |
|
50–70 |
75 |
120 |
120 |
120 |
|
70–115 |
45 |
95 |
95 |
120 |
|
115–175 |
40 |
65 |
65 |
110 |
|
175–300 |
30 |
40 |
45 |
70 |
|
300–350* |
20 |
30 |
35 |
60 |
* Agilent ovens are programmable to 450 °C, but this product was only tested to a maximum operating temperature of 350 °C. Prior to analysis, confirm the analytical column can withstand the temperatures and ramp rates you plan to use.
The GC Accelerator kit was specifically designed for Agilent 6890 and 7890 GCs equipped with mass selective detectors (MSD). As shown in Figure 3, the inserts install without interfering with a column in the front inlet position or with the connection to the MSD transfer line. Once in place, the oven inserts occupy oven volume, which reduces the volume of air that the oven needs to heat and cool. This, in turn, permits faster oven ramp rates and quicker oven cooldown times.
Figure 3: Restek GC Accelerator Oven Insert Kit.
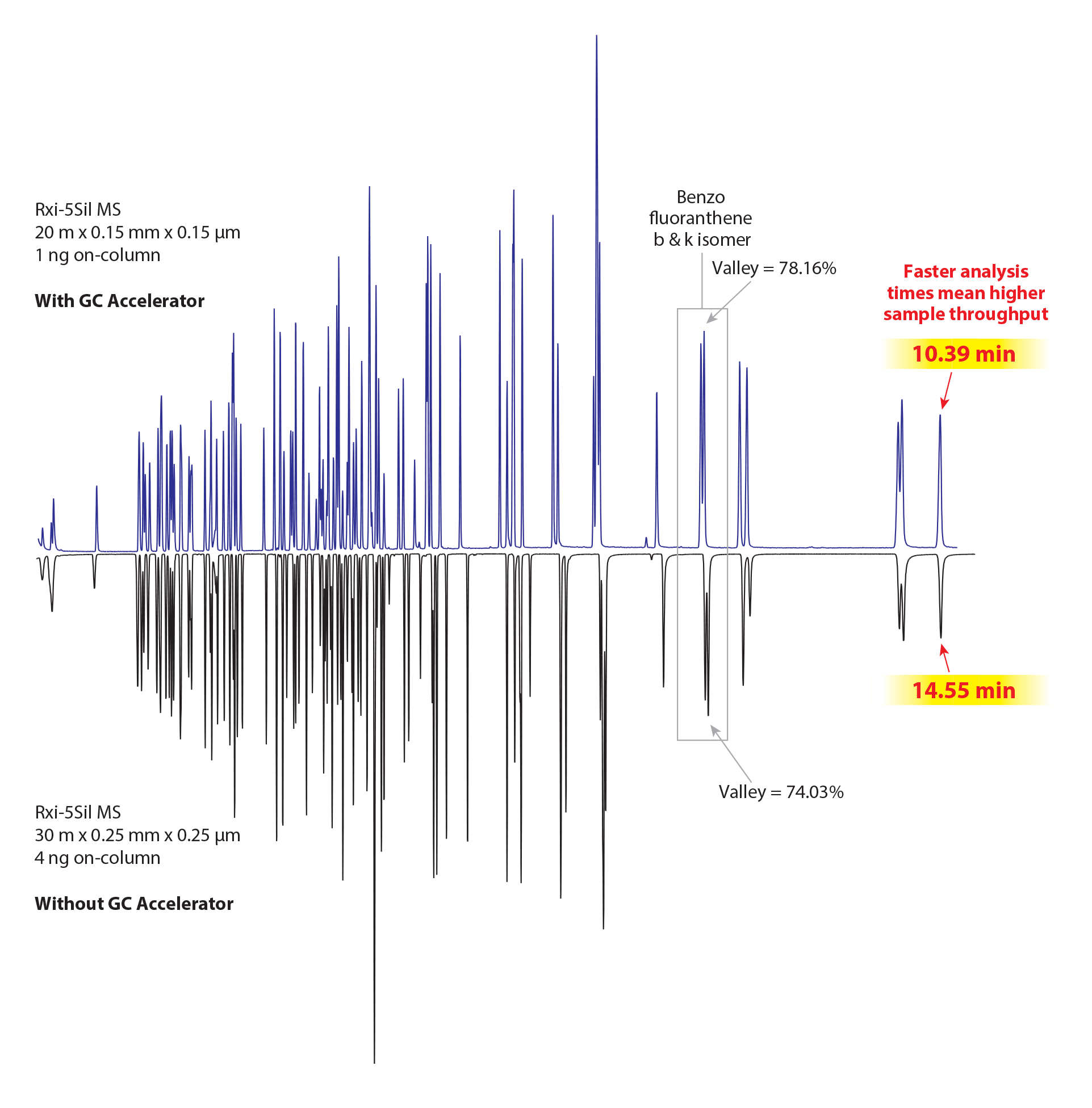
Figure 4 compares split analyses with the traditional column format (bottom) and the scaled-down column using the translated method conditions and a GC Accelerator kit (top). The chromatograms are mirrored and scaled such that the starting and ending oven temperatures are the same for both chromatograms, which allows for a direct comparison of the separations. The separation profile is practically identical, even with difficult-to-resolve critical pairs like benzo[b]fluoranthene and benzo[k]fluoranthene maintaining acceptable resolution. The last eluting compound elutes over four minutes faster with the scaled-down column and method, and the total analysis time dropped by over six minutes. The speed increase shown in the faster semivolatiles analysis would be greater still when compared to a traditional splitless injection.
Figure 4: Faster semivolatiles analysis can be obtained using a scaled-down column, properly adjusted conditions, and a GC Accelerator kit. (For reference, the instrument conditions for the top chromatogram are show below this figure, and the conditions for the bottom chromatogram are shown in Figure 1.)
| Column | Rxi-5Sil MS, 20 m, 0.15 mm ID, 0.15 µm (cat.# 43816) |
|---|---|
| Standard/Sample | 8270 MegaMix (cat.# 31850) |
| 8270 Benzidines mix (cat.# 31852) | |
| Benzoic acid (cat.# 31879) | |
| 1,4-Dioxane (cat.# 31853) | |
| Revised B/N surrogate mix (cat.# 31888) | |
| Acid surrogate mix (4/89 SOW) (cat.# 31063) | |
| Revised SV internal standard mix (cat.# 31886) | |
| Diluent: | Methylene chloride |
| Conc.: | 20 µg/mL (IS/SS 20 µg/mL) |
| Injection | |
| Inj. Vol.: | 1 µL split (split ratio 20:1) |
| Liner: | Topaz 4 mm single taper w/wool (cat.# 23303) |
| Inj. Temp.: | 275 °C |
| Split Vent Flow Rate: | 12 mL/min. |
| Carrier Gas | He, constant flow |
| Flow Rate: | 0.72 mL/min |
| Detector | MS |
|---|---|
| Mode: | Scan |
| Transfer Line Temp.: | 280 °C |
| Analyzer Type: | Quadrupole |
| Source Temp.: | 330 °C |
| Quad Temp.: | 180 °C |
| Electron Energy: | 70 eV |
| Solvent Delay Time: | 1.3 min |
| Tune Type: | DFTPP |
| Ionization Mode: | EI |
| Scan Range: | 39-550 amu |
| Scan Rate: | 9.8 scans/sec |
| Instrument | Agilent 7890B GC & 5977A MSD |
| Notes | Oven Programs Top: 70 °C (hold 0.7 min) to 285 °C at 39.8 °C/min to 305 °C at 4.3 °C/min to 320 °C at 28.5 °C/min (hold 0.7 min) Bottom: 70 °C (hold 1 min) to 285 °C at 28 °C/min to 305 °C at 3 °C/min to 320 °C at 30 °C/min (hold 1 min) |
Scaled-Down Method: Optimization of Fast Semivolatiles Analysis
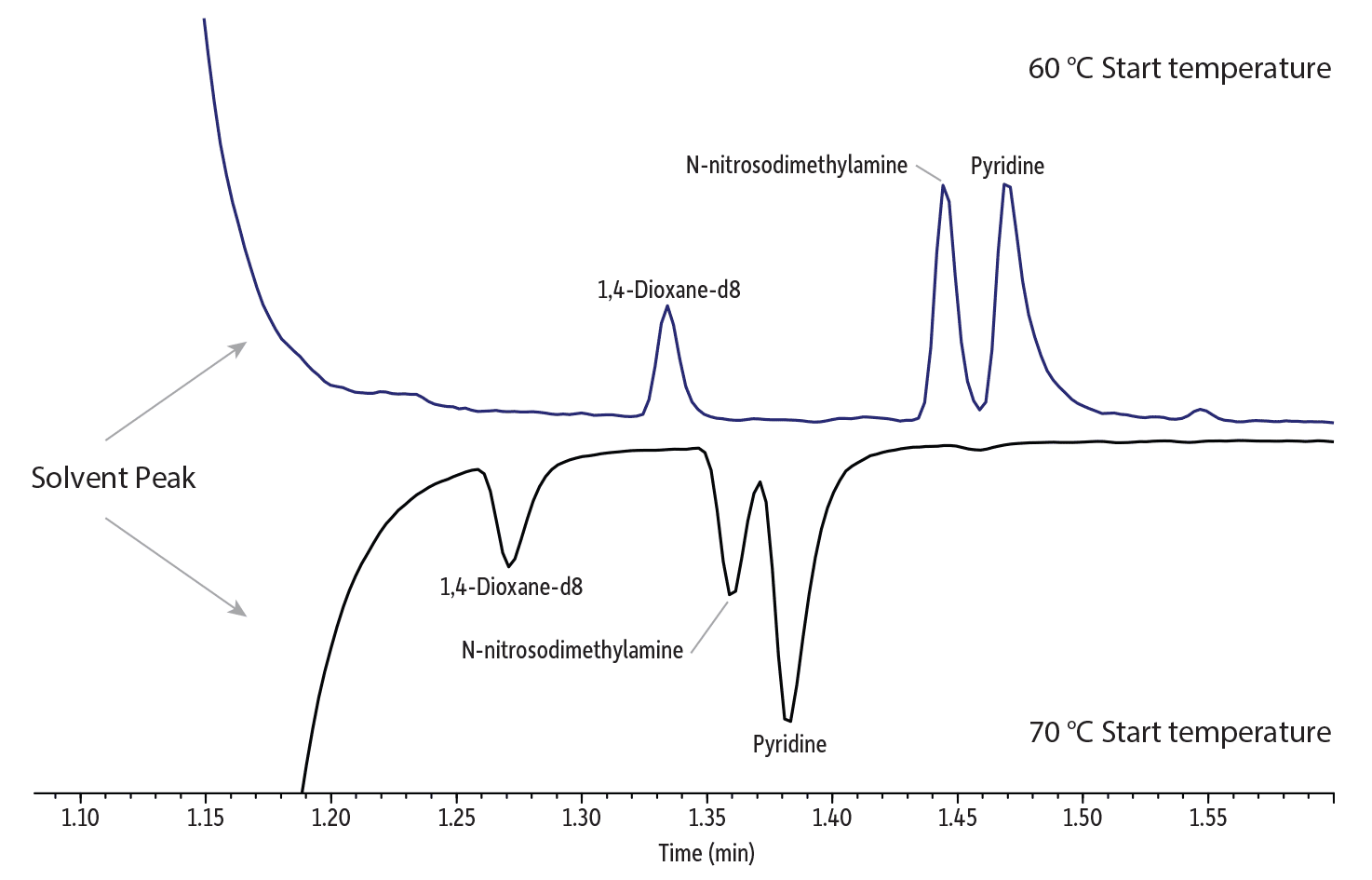
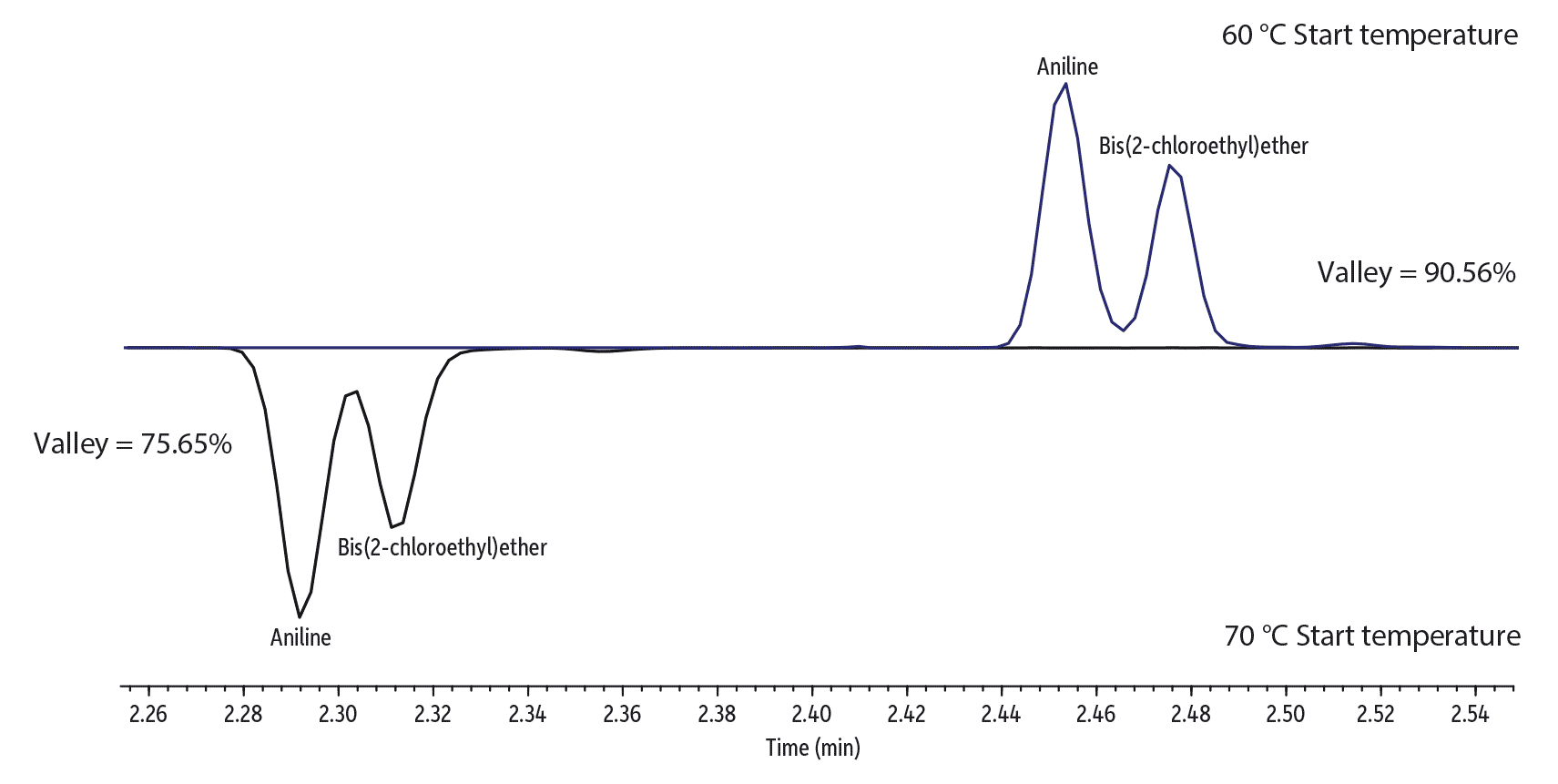
The directly translated scaled-down method required small adjustments to optimize results, particularly for early eluting compounds. Note that no changes to the oven ramp rates or the flow were necessary; the values calculated by the method translator were valid. As shown in Figure 5, even though the higher split ratio of 20:1 resulted in a very narrow sample band being transferred onto the column, resolution of the early eluting compounds was not ideal. However, by simply decreasing the initial oven temperature from 70 °C to 60 °C, additional column focusing was achieved and results were improved. Figure 5 shows how 1,4-dioxane is better resolved from the solvent peak and the separation of aniline and bis(2-chloroethyl)ether is also improved by dropping the initial oven temperature just 10 °C.
In addition to lowering the initial oven temperature, the final oven temperature was raised from the customary 320 °C to 330 °C to more effectively remove high molecular weight contamination. The time saved in the scaled-down method provided the extra time needed to reach and then cool down from the higher final oven temperature. Table III summarizes the changes made to optimize the directly translated method and Figure 6 shows the final conditions recommended for fast semivolatiles analysis using a scaled-down column, translated method, and the GC Accelerator kit.
Table III: Summary of changes to optimize the scaled-down method.
| GC Method Parameters | Traditional Split Method Conditions | Direct Method Translation from EZGC Method Translator | Final Method with Refined Initial and End Temperatures | Differences Between Direct Translation & Final Method |
|
|---|---|---|---|---|---|
| Column Flow (mL/min) | 1.2 | 0.7 | 0.7 | ||
| Oven Program | Initial (hold) | 70 °C (hold 1 min) | 70 °C (hold 0.7 min) | 60 °C (hold 0.7 min) | Lowering initial oven temp increased resolution of early-eluting compounds |
| Ramp 1 | to 285 °C @ 28 ˚C/min |
to 285 °C @ 39.8 ˚C/min | to 285 °C @ 39.8 ˚C/min | ||
| Ramp 2 | to 305 °C @ 3 °C/min |
to 305 °C @ 4.3 °C/min | to 305 °C @ 4.3 °C/min | ||
| Ramp 3 (hold) | to 320 °C @ 20 °C/min |
to 320 °C @ 28.5 °C/min | to 330 °C @ 28.5 °C/min | Raising the final oven temperature facilitates more thorough removal of high molecular weight contamination |
|
Figure 5: Lowering the translated initial oven temperature from 70 °C to 60 °C improved resolution of early eluting compounds. (For reference, the instrument conditions for the 60 °C start are shown in Figure 6 and for the 70 °C start, in the top chromatogram in Figure 4.)
Solvent Peak and 1,4-Dioxane
Aniline and Bis(2-Chloroethyl) Ether
Figure 6: Final optimized method for fast semivolatiles analysis using a scaled-down column format, translated method, and GC Accelerator kit.
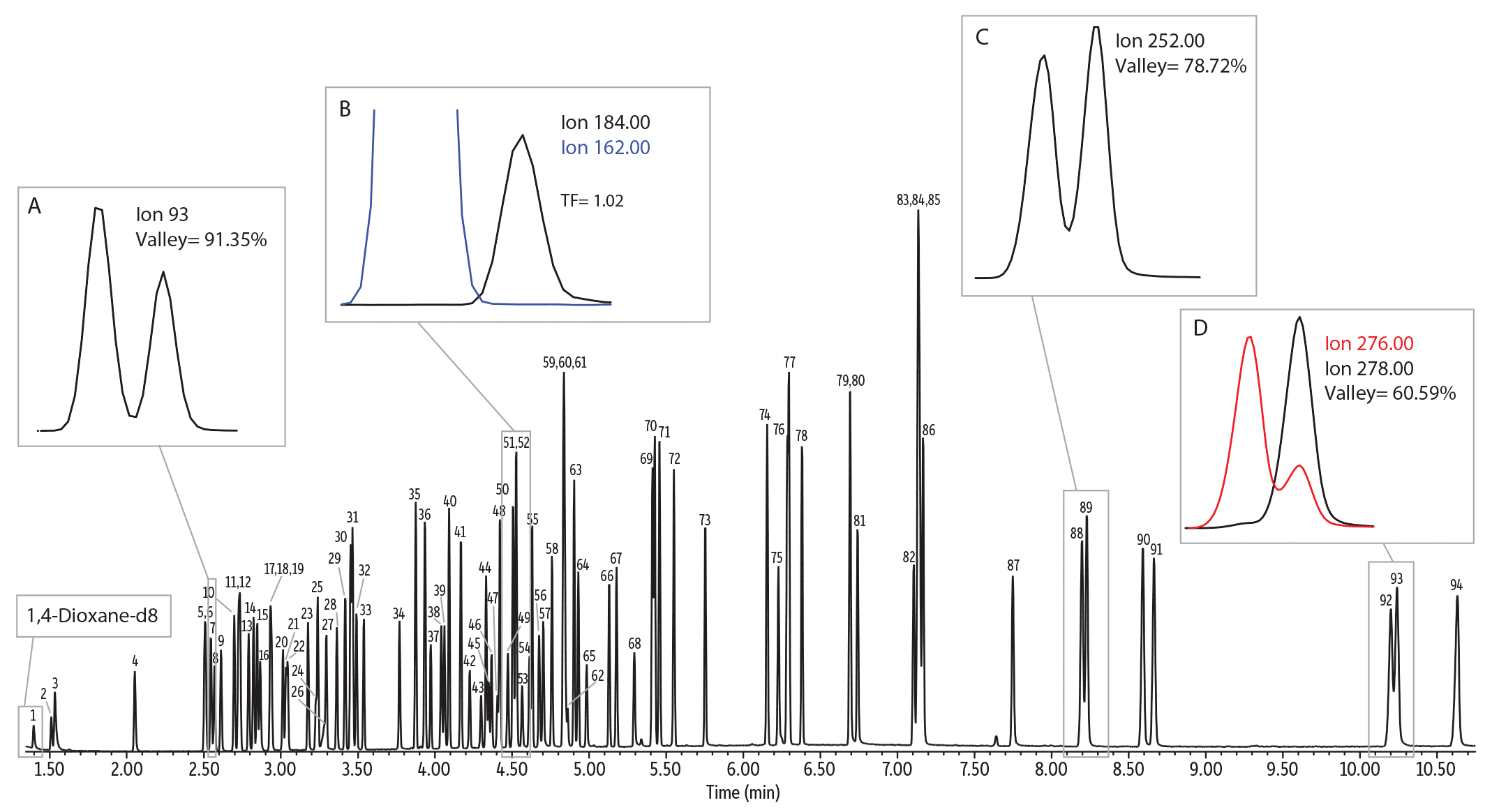
| Peaks | tR (min) | |
|---|---|---|
| 1. | 1,4-Dioxane-d8 (IS) | 1.40 |
| 2. | N-Nitrosodimethylamine | 1.51 |
| 3. | Pyridine | 1.53 |
| 4. | 2-Fluorophenol (SS) | 2.05 |
| 5. | Phenol-d6 (SS) | 2.51 |
| 6. | Phenol | 2.51 |
| 7. | Aniline | 2.55 |
| 8. | Bis(2-chloroethyl) ether | 2.57 |
| 9. | 2-Chlorophenol | 2.61 |
| 10. | 1,3-Dichlorobenzene | 2.70 |
| 11. | 1,4-Dichlorobenzene-d4 (IS) | 2.73 |
| 12. | 1,4-Dichlorobenzene | 2.74 |
| 13. | Benzyl alcohol | 2.79 |
| 14. | 1,2-Dichlorobenzene | 2.82 |
| 15. | 2-Methylphenol | 2.85 |
| 16. | Bis(2-chloroisopropyl)ether | 2.87 |
| 17. | 4-Methylphenol | 2.93 |
| 18. | 3-Methylphenol | 2.93 |
| 19. | N-Nitrosodi-N-propylamine | 2.94 |
| 20. | Hexachloroethane | 3.01 |
| 21. | Nitrobenzene-d5 (SS) | 3.03 |
| 22. | Nitrobenzene | 3.04 |
| 23. | Isophorone | 3.18 |
| 24. | 2-Nitrophenol | 3.23 |
| 25. | 2,4-Dimethylphenol | 3.24 |
| 26. | Benzoic acid | 3.28 |
| 27. | Bis(2-chloroethoxy)methane | 3.30 |
| 28. | 2,4-Dichlorophenol | 3.36 |
| 29. | 1,2,4-Trichlorobenzene | 3.42 |
| 30. | Naphthalene-d8 (IS) | 3.45 |
| 31. | Naphthalene | 3.47 |
| Peaks | tR (min) | |
|---|---|---|
| 32. | 4-Chloroaniline | 3.49 |
| 33. | Hexachlorobutadiene | 3.54 |
| 34. | 4-Chloro-3-methylphenol | 3.77 |
| 35. | 2-Methylnaphthalene | 3.87 |
| 36. | 1-Methylnaphthalene | 3.93 |
| 37. | Hexachlorocyclopentadiene | 3.97 |
| 38. | 2,4,6-Trichlorophenol | 4.04 |
| 39. | 2,4,5-Trichlorophenol | 4.06 |
| 40. | 2-Fluorobiphenyl (SS) | 4.09 |
| 41. | 2-Chloronaphthalene | 4.17 |
| 42. | 2-Nitroaniline | 4.23 |
| 43. | 1,4-Dinitrobenzene | 4.30 |
| 44. | Dimethyl phthalate | 4.33 |
| 45. | 1,3-Dinitrobenzene | 4.35 |
| 46. | 2,6-Dinitrotoluene | 4.37 |
| 47. | 1,2-Dinitrobenzene | 4.41 |
| 48. | Acenaphthylene | 4.42 |
| 49. | 3-Nitroaniline | 4.47 |
| 50. | Acenaphthene-d10 (SS) | 4.51 |
| 51. | Acenaphthene | 4.53 |
| 52. | 2,4-Dinitrophenol | 4.53 |
| 53. | 4-Nitrophenol | 4.56 |
| 54. | 2,4-Dinitrotoluene | 4.61 |
| 55. | Dibenzofuran | 4.63 |
| 56. | 2,3,5,6-Tetrachlorophenol | 4.68 |
| 57. | 2,3,4,6-Tetrachlorophenol | 4.70 |
| 58. | Diethyl phthalate | 4.76 |
| 59. | 4-Chlorophenyl phenyl ether | 4.83 |
| 60. | Fluorene | 4.84 |
| 61. | 4-Nitroaniline | 4.85 |
| 62. | 4,6-Dinitro-2-methylphenol | 4.86 |
| Peaks | tR (min) | |
|---|---|---|
| 63. | N-Nitrosodiphenylamine (as diphenylamine) | 4.90 |
| 64. | Diphenylhydrazine (as azobenzene) | 4.93 |
| 65. | 2,4,6-Tribromophenol (SS) | 4.98 |
| 66. | 4-Bromophenyl phenyl ether | 5.13 |
| 67. | Hexachlorobenzene | 5.18 |
| 68. | Pentachlorophenol | 5.29 |
| 69. | Phenanthrene-d10 (IS) | 5.41 |
| 70. | Phenanthrene | 5.43 |
| 71. | Anthracene | 5.46 |
| 72. | Carbazole | 5.55 |
| 73. | di-n-Butyl phthalate | 5.75 |
| 74. | Fluoranthene | 6.15 |
| 75. | Benzidine | 6.23 |
| 76. | Pyrene-d10 (SS) | 6.28 |
| 77. | Pyrene | 6.30 |
| 78. | p-Terphenyl-d14 (SS) | 6.38 |
| 79. | 3,3'-Dimethylbenzidine | 6.69 |
| 80. | Butyl benzyl phthalate | 6.69 |
| 81. | Bis(2-ethylhexyl) adipate | 6.74 |
| 82. | 3,3'-Dichlorobenzidine | 7.11 |
| 83. | Bis(2-ethylhexyl) phthalate | 7.13 |
| 84. | Chrysene-d12 (IS) | 7.14 |
| 85. | Benz[a]anthracene | 7.14 |
| 86. | Chrysene | 7.17 |
| 87. | Di-n-octyl phthalate | 7.75 |
| 88. | Benzo[b]fluoranthene | 8.20 |
| 89. | Benzo[k]fluoranthene | 8.23 |
| 90. | Benzo[a]pyrene | 8.59 |
| 91. | Perylene-d12 (IS) | 8.67 |
| 92. | Indeno[1,2,3-cd]pyrene | 10.20 |
| 93. | Dibenz[a,h]anthracene | 10.24 |
| 94. | Benzo[ghi]perylene | 10.63 |
| Column | Rxi-5Sil MS, 20 m, 0.15 mm ID, 0.15 µm (cat.# 43816) |
|---|---|
| Standard/Sample | 8270 MegaMix (cat.# 31850) |
| 8270 Benzidines mix (cat.# 31852) | |
| Benzoic acid (cat.# 31879) | |
| Revised B/N surrogate mix (cat.# 31888) | |
| Acid surrogate mix (4/89 SOW) (cat.# 31063) | |
| Revised SV internal standard mix (cat.# 31886) | |
| Diluent: | Methylene chloride |
| Conc.: | 20 µg/mL (IS/SS 20 µg/mL) |
| Injection | |
| Inj. Vol.: | 1 µL split (split ratio 20:1) |
| Liner: | Topaz 4 mm single taper w/wool (cat.# 23303) |
| Inj. Temp.: | 275 °C |
| Oven | |
| Oven Temp.: | 60 °C (hold 0.7 min) to 285 °C at 39.8 °C/min to 305 °C at 4.3 °C/min to 330 °C at 28.5 °C/min (hold 3.5 min) |
| Carrier Gas | He, constant flow |
| Flow Rate: | 0.72 mL/min |
| Detector | MS |
|---|---|
| Mode: | Scan |
| Transfer Line Temp.: | 280 °C |
| Analyzer Type: | Quadrupole |
| Source Temp.: | 330 °C |
| Quad Temp.: | 180 °C |
| Electron Energy: | 70 eV |
| Solvent Delay Time: | 1.3 min |
| Tune Type: | DFTPP |
| Ionization Mode: | EI |
| Scan Range: | 39-550 amu |
| Scan Rate: | 9.8 scans/sec |
| Instrument | Agilent 7890B GC & 5977A MSD |
| Notes | Analyzed using a 120 V oven equipped with the GC Accelerator kit (cat.# 23849). |
Meeting Method Requirements
All the compounds included in this fast semivolatiles analysis method evaluation met EPA 8270 method requirements of either exhibiting a %RSD of relative response factors (RRFs) of <20% or demonstrating a correlation coefficient of ≥0.99 if an alternative fit method was used. Five of the more active compounds (benzoic acid, 2,4-dinitrophenol, 4,6-dinitro-2-methylphenol, benzidine, and 3,3’-dimethylbenzidine) required the use of an inversely weighted linear curve, although the correlation coefficient in those cases met the method requirements.
In Summary
By applying the principals of column and method scale-down, with the assistance of the GC Accelerator kit to provide the added boost to your oven ramp rates, the analysis of even complex samples can be successfully translated to a much faster method while maintaining the same separation. In the example shown here, scaling down EPA Method 8270 resulted in much faster semivolatiles analysis times that will allow labs to increase sample throughput or process more rush samples.